
Zyvex
is pleased to offer our first development-version CAD tool to the world.
This is now available for free download from
the Zyvex web site.
|
Original 3D design from SolidWorks |
Decomposed into atoms by DiamondCAD |
This software was originally based on some programming done by Will Ware (Algorithmic generation of molecular structures). It was modified, extended, and interfaced to HyperChem by Chris Phoenix (http://www.best.com/~cphoenix) during his visit to Zyvex in early 1998. We wish to thank both Will and Chris for their willingness and desire to freely distribute their software. The newest version of this code was written in Visual Basic by John Michelsen while he worked at Zyvex.
We are using this to explore atomic decomposition of solid 3D structures, and welcome involvement of others in this project. We are giving away the source to this software as an experiment in open software development. We understand that reliance on commercial software like SolidWorks and HyperChem may "taint" the project for some, but believe it is overly pure to insist on recreating hundreds of manyears of work before even getting started. Better to develop only the unique glue, in our opinion.
If you would like a sponsored trip to Zyvex to work on this project, send us an email with your resume and ideas for carrying this project forward. We are interested in collaborations ranging from occasional contributors to consultants to full-time staff, depending on your level of interest and ability. This project may join with the OpenChem effort, depending on how that work goes.
DiamondCAD traverses a SolidWorks assembly, and for each component, traverses its feature list, filling cylinders, rectangular solids and helices with diamond. It then adds hydrogens to unterminated valences and reconstructs the resulting surface by searching for hydrogens that are too close to each other and deleting them, adding a new bond between their carbons. DiamondCAD can output the entire PDB file to HyperChem, or just the outer hydrogens of components to get by HyperChem's maximum atom limit of 91000 atoms, and to speed up visualization generally.
The above images show an assembly as generated in SolidWorks and decomposed into atoms for HyperChem. The following images show a concept for a feedstock acquisition system in SolidWorks. By clicking on the images, you can see the resulting molecules for HyperChem in stick and CPK renderings. Further assembler files output by DiamondCAD can be seen here.
|
Feedstock
acquisition 1 |
Feedstock
acquisition 2 |
SORRY: links are currently broken, with no ETA on restoring them.
DiamondCAD 0.3 is available now from our web site. To download the latest version, click here.
DiamondCAD 0.2 (independent from both HyperChem and SolidWorks) is available now here.
DiamondCAD 0.1 is an extension to HyperChem 5 and is available here in Zip format. It allows more shape types, filling in SiC, silicon, BN, AlN, and SiO2 lattices, and viewing atom by atom construction on diamond 100 and 110 planes.
Here is the executable for Ralph Merkle's tube.c interfaced to HyperChem. The following image is from one of his example inputs.
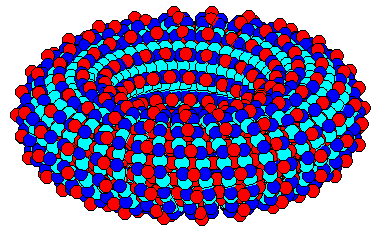
After investigating several commercial solid modeling packages, Zyvex chose SolidWorks 98 as its CAD system of choice, not least because of its parametric capabilities and its user API. Taking advantage of the latter feature, it was not too difficult to put together a translator from SolidWorks parts to DCAD 0.2 files. This works by the admittedly slow method of intersecting cubes with SW98 parts, and writing them to a file if they are found to be inside of it. After running it overnight on a scaled down canted rotary joint, the following was produced:
Above figures are CPK and stick renderings of a SolidWorks part run through SW2MolEd and written out as a PDB file. This and other PDB files that can be viewed by any number of chemistry programs can be downloaded here.
For those of you with access to SolidWorks, here is the Visual Basic source and executable for the program. A rough sketch of an assembler replicating can be seen here and can be viewed in 3D by downloading the following files and the free SolidWorks viewer.
A simulation of a bearing (size warning: this GIF is about 2.6MB!) generated by DiamondCad 0.3. The simulation is driven by the same software as below.
We're also working on a C port of Don Brenner's Reactive Hydrocarbon Code to add the ability to simulate realistic mechanosynthetic construction reactions and larger scale bond breaking. This code scales linearly with system mass instead of the higher orders of semi-empirical calculations. In the meantime, here is a Fortran executable allowing interactive mechanochemistry simulations to be performed with HyperChem 5.1 or HyperChem's free demo program. The demo program seems to open up very slowly on Win98 and says your time is up after 10 minutes or so. I haven't tried it on Win95, but it is a very pretty program. Look at the Readme file in the zip archive for some installation and use instructions.
Here is some VB code that allows mechanochemistry simulations to be performed with HyperChem 5's own mehcanics and semi-empirical potentials. On input of two molecules you would like to translate together, slide relative to one another, or rotate, it asks for slabs of atoms on each to impart forces on. These are easily selected by dragging a box around them with HyperChem's selection tool. Depending on the movement choices you selected, it then prompts for a slide vector or rotation axis. If the active force field is a semi-empirical or ab-initio potential, clicking on "Do approaches" will rather realistically bring the molecules together and apart the specified distance, in specified increments, with the specified number of dynamics time steps between increments. If the active force field is a mechanics module, frictional analysis can be performed. Using the PM3 semi-empirical potential on model compounds resembling a diamond STM tip and the diamond(100) surface resulted in the following animated GIFs (generated from many output .bmp files by "Graphic Converter" for the Mac. You may need to save the GIFs to disk to get them to play fast enough):
H abstraction with a cyclopropyl carbene
Failed abstraction attempt with cyclopropene carbene
Unreactive crash of saturated molecules
Unreactive crash of unsaturated molecules
Reversible carbon atom transfer, (larger model compounds)
Carbon transfer next to an adatom site, (failed attempt with larger model compounds)
Carbon dimer transfer to a groove site
Reversible nitrogen atom transfer
Failed oxygen atom transfer
Reversible phosphorus atom transfer
Reversible silicon atom transfer
Reversible sulfur atom transfer
Failed flourine atom transfer
Reversible chlorine atom transfer
Reversible aluminum atom transfer between model C(100) molecules
Reversible carbon atom transfer between model C(100) and graphite compounds
Failed carbon atom transfer to extend a graphene sheet
Carbon atom transfer to a carbyne radical.
Reversible silicon atom transfer between model Si(100) compounds
Reversible hydrogen atom transfer, (larger model compounds )
Hydrogen deposition from R3SiH group
Acetylene abstraction from a dimer
Acetylene deposition onto a dimer, (larger model compounds)
Acetylene deposition across dimers, (larger model compounds)
Hydrogen abstraction with a carbyne radical
Refreshing of a carbyne radical, SiR3 catalyst
Failed carbon deposition by the twist method
If
you'd like to make modifications and pass them back, we'd be happy to post
them (and use them). We also have lots of ideas for future directions; email
Zyvex if you'd like to get onboard.
Trademarks used herein are the property of their respective holders, and may be registered in some domains. Zyvex and DiamondCAD are trademarks of Zyvex LLC.